Last Updated on 2024-08-11 by Clay
Singular Value Decomposition (SVD) is a method for decomposing a matrix into three matrices, revealing the rank, data dimensions, and key directions of the original matrix. It is often used in dimensionality reduction, compression, and structural analysis.
Let be a matrix of size
. The SVD can be written as:
Here, has dimensions
,
has dimensions
, and
has dimensions
. Additionally:
is an orthogonal matrix, meaning
, and represents the left singular vectors of
.
is also an orthogonal matrix, meaning
, and represents the right singular vectors of
.
is a diagonal matrix (though not necessarily square), with non-zero values only on the diagonal. These diagonal values are the singular values of
.

In the figure above, represents the rank of the matrix
, which is the maximum number of linearly independent rows or columns in
. It also corresponds to the number of non-zero singular values in the
matrix, indicating the independent information contained within
.
On the other hand, also influences the effective range of the decomposed matrices
,
, and
. Generally, when
and
, we can reduce the size of the decomposed matrices by removing certain rows or columns from
and
, thereby saving storage space. This approach is common in data compression methods, such as PCA.
How to Compute SVD
To compute SVD, follow these steps:
- Compute
and
- Solve for
and
- Solve for
- Assemble
,
,
Along the way, I’ll verify the calculations using Python.
Let’s assume we have the following matrix :
We will attempt to decompose it using SVD.
1. Compute  and
and 
Next, we calculate and
:
Let’s verify these calculations using Python:
import numpy as np
A = np.array(
[
[3, 2],
[2, 3],
[1, 1],
],
)
# A^T A
At_A = np.dot(A.T, A)
print("A^T A =\n", At_A)
print("\n")
# A A^T
A_At = np.dot(A, A.T)
print("A A^T =\n", A_At)
Output:
A^T A = [[14 13] [13 14]] A A^T = [[13 12 5] [12 13 5] [ 5 5 2]]
2. Solve for 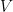 and
and 
Compute Eigenvalues
First, we solve for matrix and the singular value matrix
by performing eigenvalue decomposition on the matrix
.
The eigenvalues satisfy the following characteristic equation:
We can then compute the equation for solving :
Finally, solving the equation gives:
Compute Singular Values
The singular values are the square roots of the eigenvalues, which we use to construct the matrix:
So the matrix is:
Compute Eigenvectors
Next, we can solve for the eigenvectors using the following equation:
When :
This gives:
After normalization, the eigenvector is:
When :
Similarly, after normalization:
Finally, we obtain as:
Let’s verify this using Python:
# Calculate eigenvalues and eigenvectors of `A^T A`
eigvals_AtA, eigvecs_AtA = np.linalg.eig(At_A)
# Caculate singular values
singular_values = np.sqrt(eigvals_AtA)
# Arrange eigenvalues and corresponding eigenvectors in descending order (make sure they are consistent with np.svd)
idx = eigvals_AtA.argsort()[::-1]
eigvals_AtA = eigvals_AtA[idx]
eigvecs_AtA = eigvecs_AtA[:, idx]
print("The Eigenvalues of `A^T A`:\n", eigvals_AtA)
print("\n")
print("The Eigenvectors of `A^T A` (V):\n", V)
print("\n")
print("Singular Values (Σ):\n", singular_values)
Output:
The Eigenvalues of `A^T A`: [27. 1.] The Eigenvectors of `A^T A` (V): [[ 0.70710678 -0.70710678] [ 0.70710678 0.70710678]] Singular Values (Σ): [5.19615242 1. ]
3. Solve for 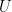
Solving for follows essentially the same steps as before. However, since the dimensions of
are larger, the calculations are a bit more involved. Here, I’ll let the code do the heavy lifting for me.
eigvals_AAt, eigvecs_AAt = np.linalg.eig(A_At)
print("The Eigenvalues of `A A^T`:\n", eigvals_AAt)
print("\n")
print("The Eigenvectors of `A A^T` (U):\n", eigvecs_AAt)
Output:
The Eigenvalues of `A A^T`: [ 2.70000000e+01 1.00000000e+00 -1.33858608e-16] The Eigenvectors of `A A^T` (U): [[ 6.80413817e-01 7.07106781e-01 -1.92450090e-01] [ 6.80413817e-01 -7.07106781e-01 -1.92450090e-01] [ 2.72165527e-01 2.07171358e-16 9.62250449e-01]]
Step 4. Assemble 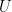 ,
,  ,
, 
Before we start assembling ,
, and
, we need to ensure that the directions of
and
are consistent. This is an important step in the SVD decomposition process.
This is because when calculating eigenvectors, the direction of the eigenvector is arbitrary. In other words, if is an eigenvector, then
is also an eigenvector.
# Get (m, n)
m, n = A.shape
min_dim = min(m, n)
# Get Σ
Sigma = np.zeros((m, n))
Sigma[:min_dim, :min_dim] = np.diag(singular_values)
# Get U, V
U = eigvecs_AAt
V = eigvecs_AtA
# Check the direction
for i in range(min_dim):
if np.dot(A, V[:, i])[0] * U[0, i] < 0:
U[:, i] = -U[:, i]
# Reconstruction
A_reconstructed = np.dot(np.dot(U, Sigma), V.T)
print("A_reconstructed:\n", A_reconstructed)
Output:
A_reconstructed: [[3. 2.] [2. 3.] [1. 1.]]
Conclusion
This article briefly discusses the process of Singular Value Decomposition (SVD), explaining how to compute ,
, and
matrices, with a focus on the importance of ensuring directional consistency between
and
.
This process not only demonstrates the mathematical principles behind SVD but also highlights the practical considerations during computation.
Understanding these details is crucial for correctly using this powerful tool in practice. Whether in data compression, dimensionality reduction, image processing, or recommendation systems, a deep understanding of SVD’s working principles can help us better apply and optimize related algorithms.
In AI-related research, the more we seek to reduce storage space, the more often we explore the applications of SVD. The importance of SVD cannot be overstated.